The Public Intellectual Property Resource for Agriculture (PIPRA) was founded in 2004 by the
Rockefeller Foundation.
Committed to participating and promoting strategies to manage public-sector intellectual property to support US and developing-country agriculture.
45 institutional members in 13 countries.
Among PIPRA’s core activities are:
- Encouraging public institutions to make informed decisions about where and when to patent
- Encouraging humanitarian exemptions in license language
- Developing a clearinghouse of public IP information and analytical resources
- Developing consolidated technology packages, or patent pools, particularly in the area of enabling technologies for plant transformation.
PIPRA created a public institution patent clearinghouse, an online, publicly accessible database that consolidated thousands of patents owned by universities and public sector research organizations.
SELECTABLE MARKERS
Notably, although there are over fifty plant selectable marker systems used in plant genetic engineering, only three used in over 90% of the scientific publications: antibiotic resistance
to kanamycin (nptII, neomycin phosphotransferase II) and hygromycin (aphIV, hygromycin phosphotrasferase), and herbicide tolerance to phosphinothricin, the active ingredient in BASTA
(Bayer CropScience, Hawthorn, Victoria, Australia) (bar or pat, phosphinothricin N-acetyltransferase).
The prevalent use of nptII in commercial crops is attributed in part to the selection system’s
effectiveness and deregulated status in the US and numerous other international government agencies. The selectable marker (nptII) is listed by the United States Food and Drug Administration
as generally recognized as safe (GRAS).
PROMOTERS
PIPRA designed and developed plant transformation DNA plasmids with maximum FTO using the nptII gene as selection system driven by a promoter different than the CaMV 35S and 19S.
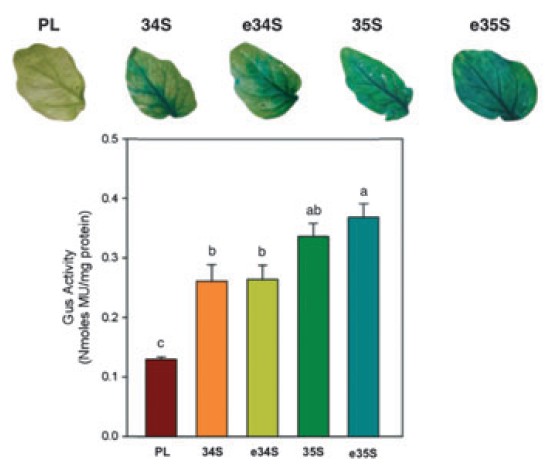
PIPRA chose the University of California’s FMV34S promoter because of its legal access and
comparable expression profile as the CaMV35S.
Figwort mosaic virus, 34S Promoter Expression
TRANSFORMATION
Single T-DNA binary vectors are the predominant option when creating GE crops. The 1T-DNA construct facilitates selection of genetically modified cells by co-integration of the selection
marker and GOI. PIPRA’s 1T-DNA vector is suitable for both research applications of new trait genes as well as engineering crops with commercial application.
To minimize potential issues due to MTA, pPIPRA T-DNA binary vectors, pPIPRA560 and pPIPRA561 include de novo synthesized bacterial origins of replication (ColE1 for E. coli and
pVS1 for propagation in Agrobacterium or Rhizobium) and the kanamycin-resistant bacteria selection marker aadA for maintenance in bacterial cells.
Given the modular design of PIPRA’s vectors, it is possible to develop the transformation vector with nonpatented components.
Current versions of the vectors are offered with promoter elements that are proprietary but for which rights can be obtained.
