The PVPO will accept differences using molecular techniques (DNA fingerprinting) only if:
- The molecular marker locus is publicly disclosed, cited, clearly identified, and detectable by a third party
- Photographic copies contain sufficient resolution of scientific publishable quality gels or other molecular data with sufficient resolution and labeling to resolve the individual data in question are provided
- The molecular data is treated the same as other methods used to establish distinctness (morphology and physiology)
- The molecular date meets the quality controls in place for appropriate supporting evidence. For example, when used to establish distinctness, the molecular data must prove that the difference is present in all individuals of the varieties and can be relied upon to prove the distinctness to anyone who performs the tests
Specifically, in the case of:
SNPs: the locus is defined by the SNP sequence showing polymorphisms
SSRs: the locus may be defined by primer pairs or sequence
AFLPs: the locus is defined by primer pairs
RAPDs: the locus is defined by primer pairs
Advantages
- Differentiates a new variety from a few older varieties that are retrieved by the computer search faster than doing grow-out trials to establish morphological differences
- Establishes that the application variety is different from the most similar comparison varieties, particularly when a gene has been inserted in the new variety and its presence or absence makes the variety distinct from other varieties
Percentage of PVPs with DNA Profiling
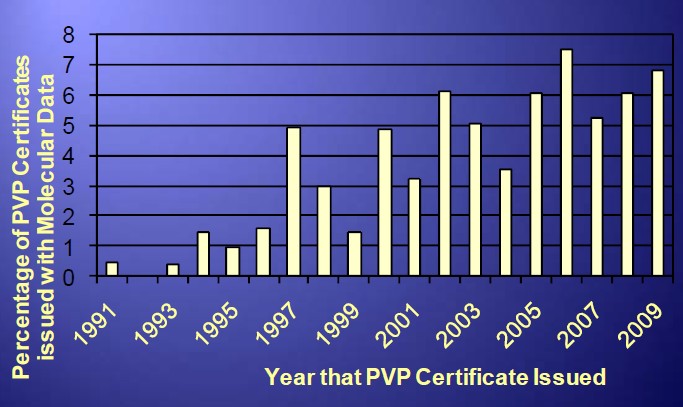
Potential Disadvantages
- Inability to distinguish a new variety from all previously existing varieties (ie older varieties with no molecular profiles)
- Insufficiently uniform and stable